Usage examples¶
Procedures and scenarios described here present few typical use cases in
manipulation of the neuron morphology reconstructions supported by the
module treem. The module should be installed and accessible. Check
that the importing works and returns no error:
python3 -c "import treem"
A Python version 3.7 or greater is assumed here. Make sure the files from
tests/data/ in the module’s source tree are copied to the working
directory.
API use cases¶
Loading the reconstruction
>>> from treem import Morph
>>> m = Morph('pass_simple_branch.swc')
>>> m.data
array([[ 1. , 1. , 0. , 0. , 0. , 1. , -1. ],
[ 2. , 3. , 1. , 1. , 0. , 0.2, 1. ],
[ 3. , 3. , 2. , 2. , 0. , 0.2, 2. ],
[ 4. , 3. , 1. , 3. , 0. , 0.2, 3. ],
[ 5. , 3. , 0. , 4. , 0. , 0.02, 4. ],
[ 6. , 3. , -1. , 5. , 0. , 0.2, 5. ],
[ 7. , 3. , -2. , 6. , 0. , 0.2, 6. ],
[ 8. , 3. , 3. , 3. , 0. , 0.2, 3. ],
[ 9. , 3. , 4. , 4. , 0. , 0.2, 8. ],
[10. , 3. , 3. , 5. , 0. , 0.2, 9. ],
[11. , 3. , 2. , 6. , 0. , 0.2, 10. ],
[12. , 3. , 5. , 5. , 0. , 0.2, 9. ],
[13. , 3. , 6. , 6. , 0. , 0.2, 12. ]])
Traversing the tree
Traverse the tree starting from the root node.
>>> start_node = m.root
>>> [node.ident() for node in start_node.walk()]
[1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13]
Traverse the tree starting from the node with ID 8.
>>> start_node = m.node(8)
>>> [node.ident() for node in start_node.walk()]
[8, 9, 10, 11, 12, 13]
By default the tree is traversed in the descending order. In the
above example a sub-tree (a branch) which starts at node with ID 8
is iterated. To walk the tree in ascending order, set the reverse
argument of treem.Node.walk().
>>> [node.ident() for node in start_node.walk(reverse=True)]
[8, 3, 2, 1]
Note that the full tree is not traversed when reverse is set: the
iterations stop as soon as the root node is reached (ID 1).
The data stored at each node can be accessed during the tree
traversal. Calculate the total length of the branch from the
start_node:
>>> sum(node.length() for node in start_node.walk())
8.485281374238571
An alternative way of traversing the tree is iterating through the sections (in descending order):
>>> for sec in start_node.sections():
>>> print([node.ident() for node in sec])
[8, 9]
[10, 11]
[12, 13]
The total length of the branch calculated this way will be the same:
>>> sum(m.length(sec) for sec in start_node.sections())
8.485281374238571
CLI use cases¶
Checking the input file
Check the input SWC file for structural consistency and compliance
with the format requirements implied by the module treem. For
example, records in the file fail_unordered.swc are not in the proper
order so the check command reports problems:
swc check fail_unordered.swc
node1_not_id1: 2
node1_has_parent: 1
node1_not_soma: 3
not_valid_parent_ids: 1
Converting the input file
Try to convert the input file to the compliant format. If there are no critical mistakes in the file structure, the conversion completes without errors:
swc convert fail_unordered.swc -o out.swc
converted 7%
converted 15%
converted 23%
converted 30%
converted 38%
converted 46%
converted 53%
converted 61%
converted 69%
converted 76%
converted 84%
converted 92%
converted 100%
Check that the output file is fine:
swc check out.swc
Displaying the morphology
Command view displays the structure of the morphology reconstruction. It only shows the center line of the reconstructed segments without their diameters:
swc view pass_nmo_1.swc
The root node is shown with the bold black dot; the soma points are shown with semitransparent spherical markers; colored lines correspond to the neurites of different types.
To display multiple cells, change the color mode to colorize individual cells:
swc view -c cells pass_mouselight_1.swc pass_mouselight_2.swc
Measuring morphometry of the reconstruction
Command measure prints out basic morphometric features of the reconstruction:
swc measure pass_nmo_1.swc
pass_nmo_1
apic area 12584.5
apic breadth 32
apic contrac 0.932844
apic degree 2
apic diam 0.816706
apic length 5690.72
apic nbranch 31
apic nstem 1
apic nterm 32
[...]
dend xdim 537.78
dend ydim 332.53
dend zdim 158.9
soma area 3137.66
soma diam 18.246
soma volume 9541.64
soma xroot 0
soma yroot 0
soma zroot 0
Locating single nodes
Command find helps to locate single nodes satisfying multiple search criteria. For example, to find a node within the dendrites (point type 3) with the diameter less than 0.1 micrometer, run the following:
swc find pass_simple_branch.swc -p 3 -d 0.1 --comp lt
5
The following command searches for the nodes of topological order 1 (which belong to the primary neurite sections):
swc find pass_simple_branch.swc -e 1
And this bash command displays the terminal sections of the dendrites:
swc view pass_simple_branch.swc -b `swc find pass_simple_branch.swc -p 3 -b 1 --sec`
Repairing damaged reconstructions
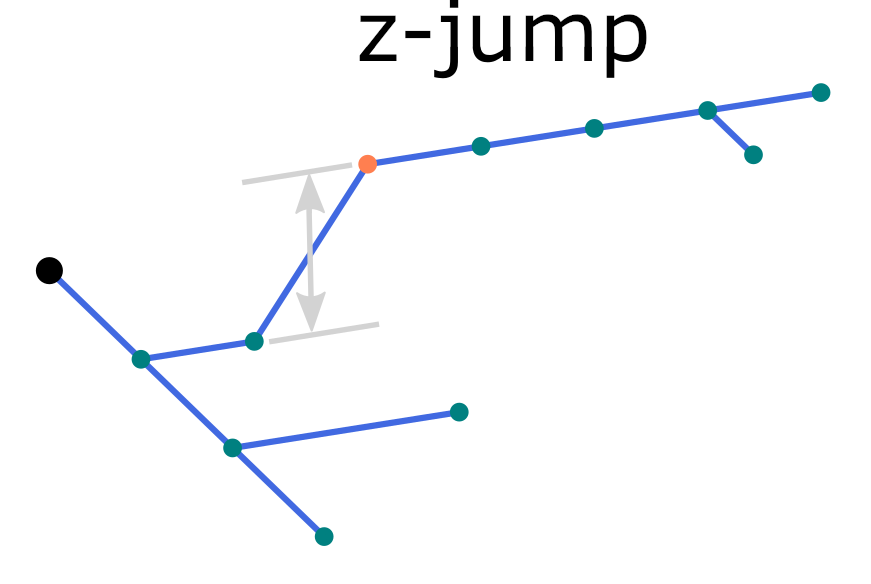
A common reconstruction error is so called z-jump when a part of the neurite gets shifted along the z-axis by few micrometers.
To locate z-jumps greater than 10 micrometers, run the command find:
swc find pass_zjump.swc -z 10
4
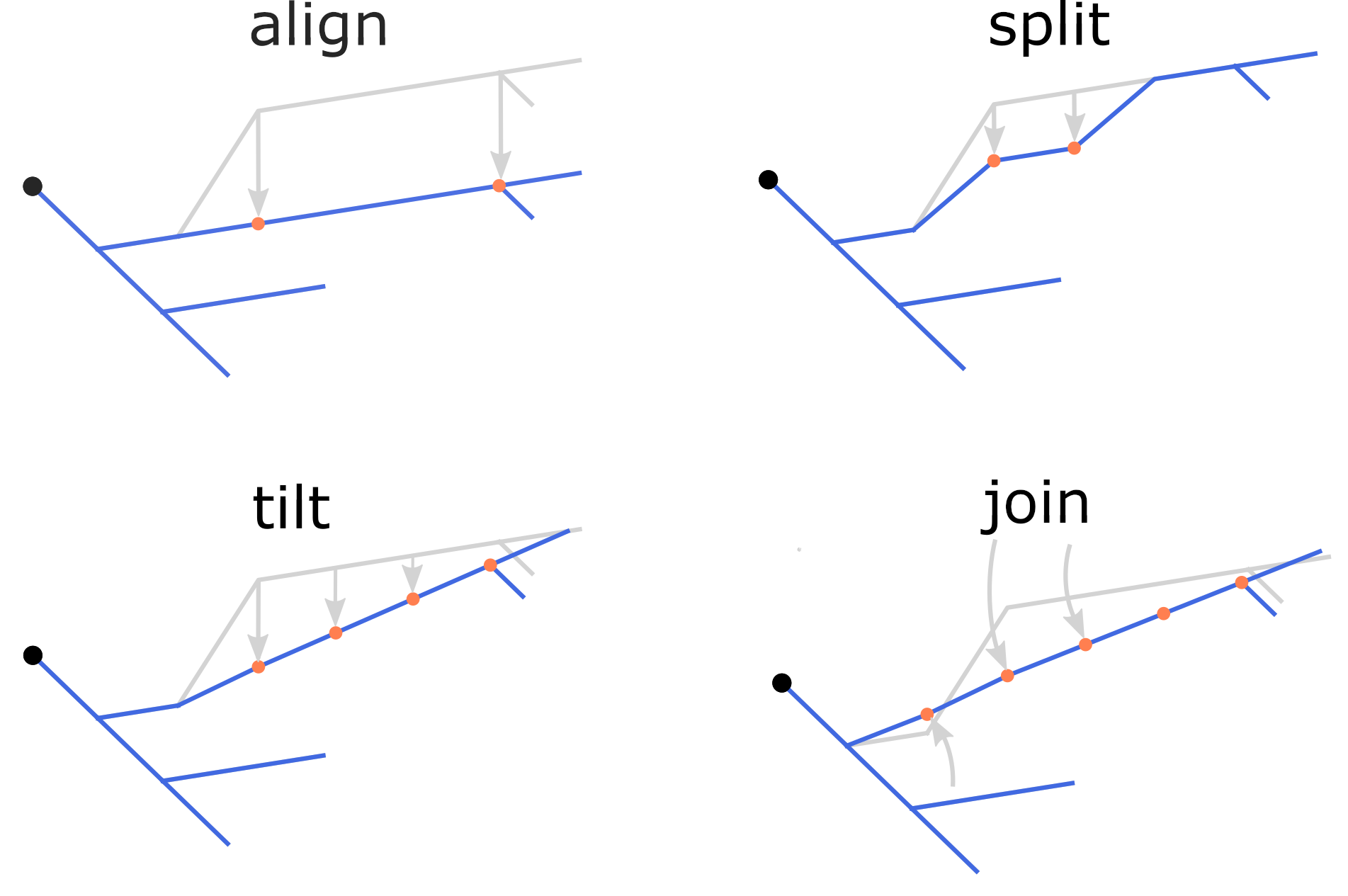
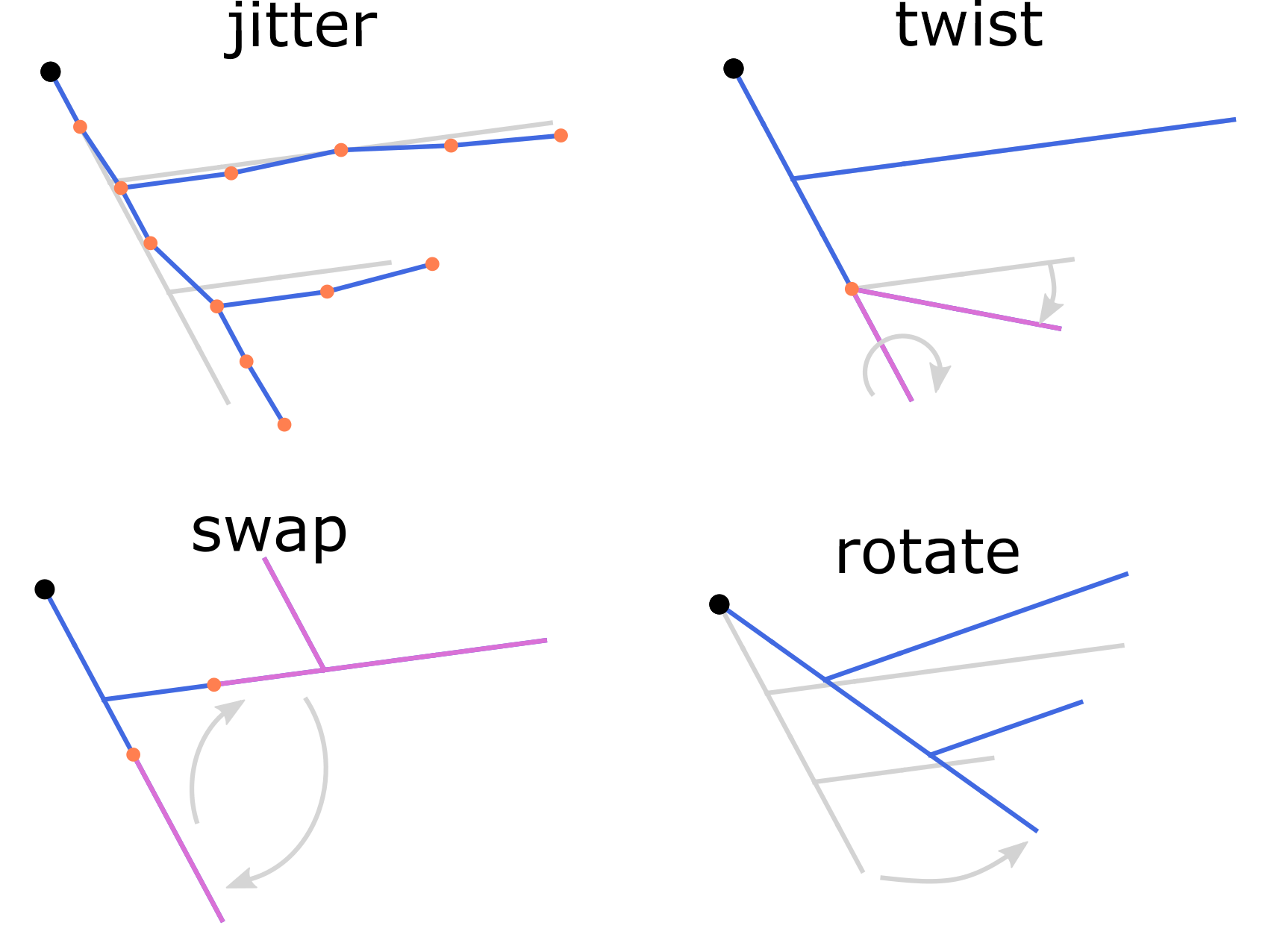
Possible z-jumps can be eliminated by the repair command using one of the four methods, align, split, tilt or join (defaults to align), as illustrated in the figure. Repair z-jumps:
swc repair pass_zjump.swc --zjump join -z `swc find pass_zjump.swc -z 10`
Experimental protocols of the slice preparation may lead to the tissue
shrinkage. Due to the shrinkage, morphology reconstructions become smaller
with increased contraction of the neurites in comparison to in vivo
conditions. To compensate for the shrinkage, various options of the
commands repair and modify can be used. See options -s, -t
and -m of the command modify for scaling, stretching and smoothing,
respectively, and options -k and -kxy of the command repair
for the shrinkage correction in z and (x, y) directions, respectively.
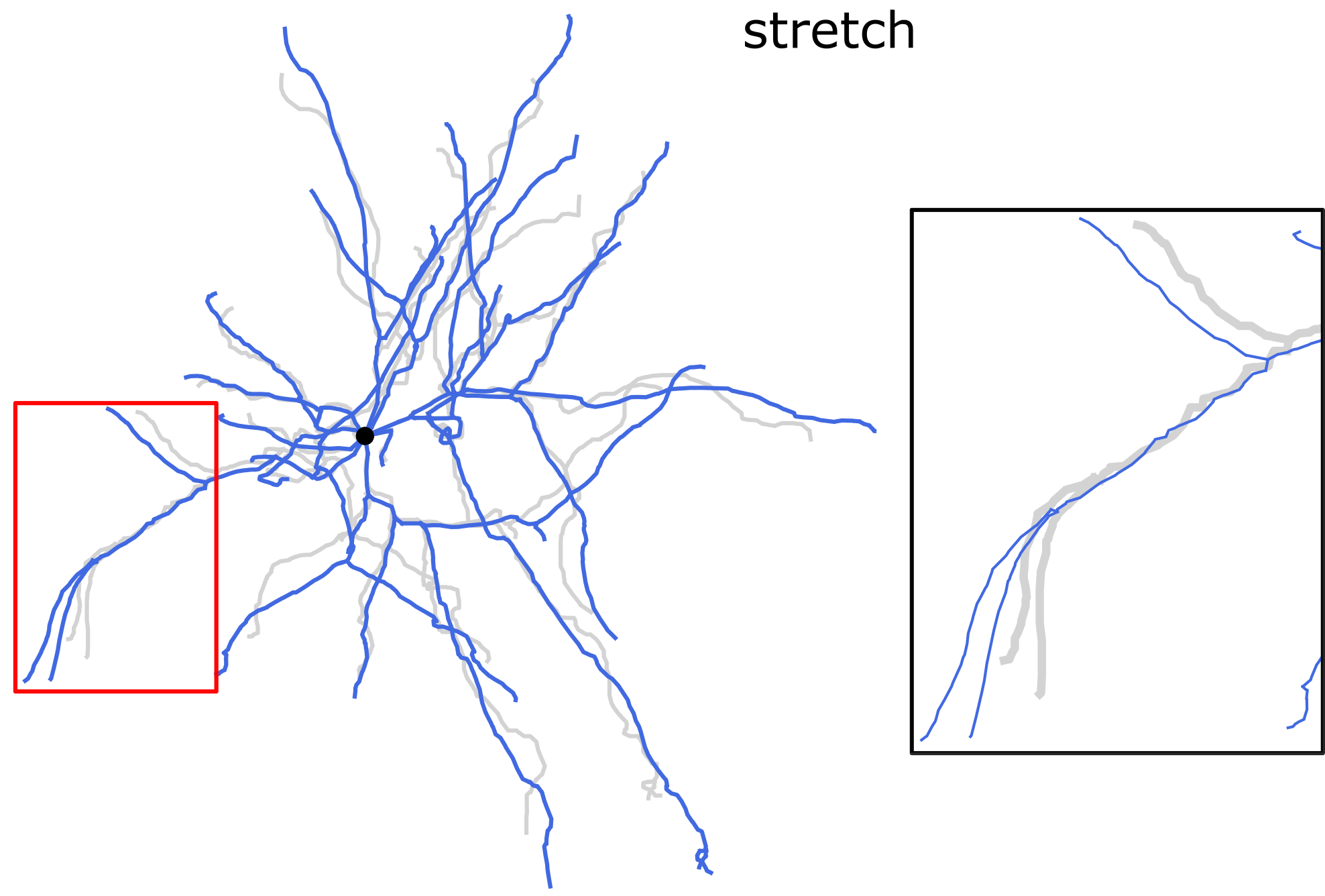
Stretching the dendrites along the direction of each dendritic section. Length-preserving operation.
Morphological reconstructions of the neurons located close to the surface of the preparation are often incomplete, missing the neurites cut by the tissue slicing. The cut points of the dendrites can be located using command find:
swc find pass_nmo_2_cut.swc -c 10 -p 3
322 341 547 1167
Here we assume the cut points are within 10 micrometers from the top surface
of the slice along the z-axis. To invert the surface orientation,
add the option --bottom-up.
Inspect the cut points in the projection and note the ID numbers for repair:
swc view pass_nmo_2_cut.swc -p 3 -j xz --show-id -m `swc find pass_nmo_2_cut.swc -c 10 -p 3`
If all cut points are located correctly, pass them all to the repair procedure:
swc repair pass_nmo_2_cut.swc -c `swc find pass_nmo_2_cut.swc -c 10 -p 3`
Alternatively, pass selected IDs to the option -c of the command repair.
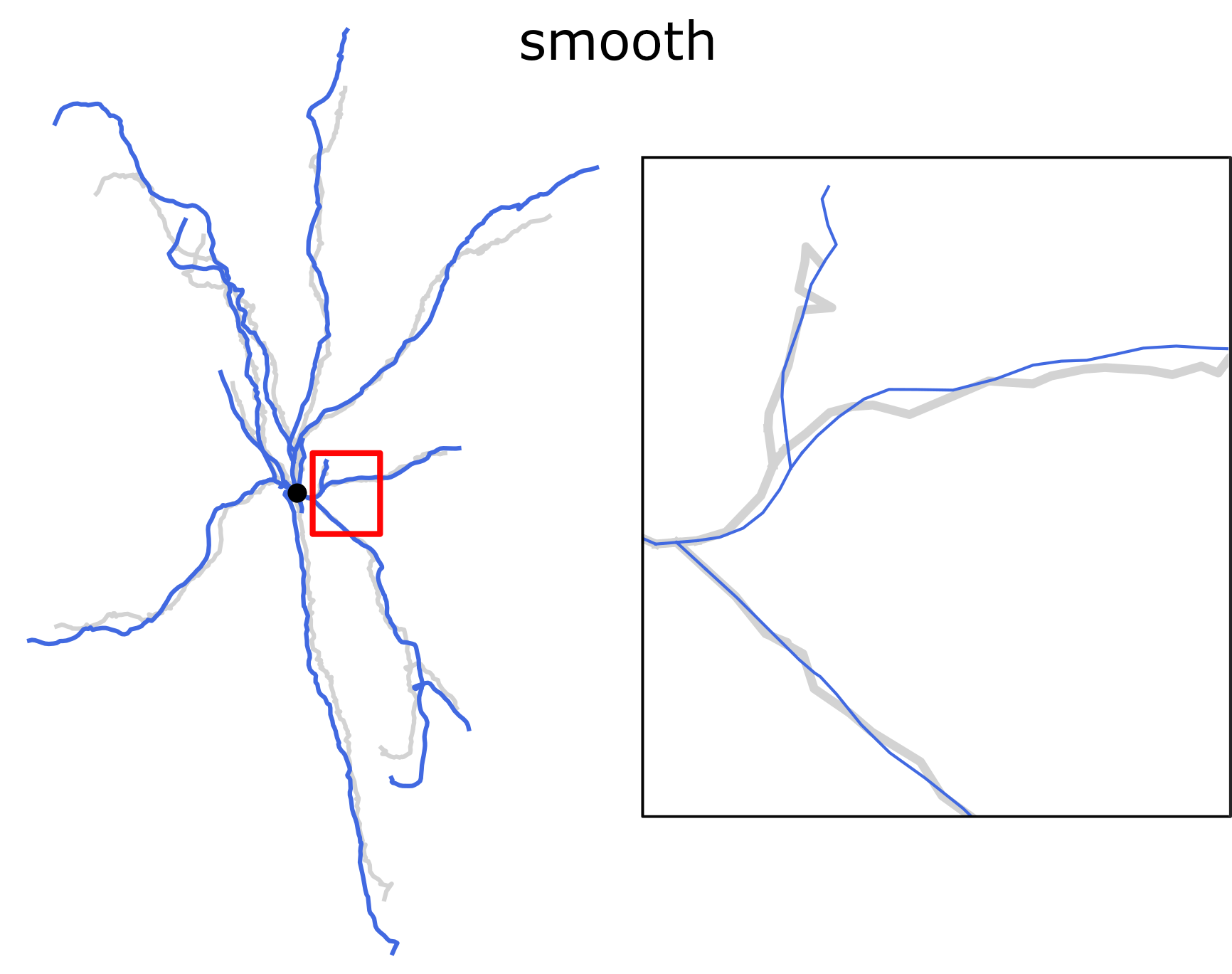
The repaired reconstruction rep.swc can be compared to the original
using the command view with -c shadow option:
swc view pass_nmo_2_cut.swc rep.swc -p 3 -c shadow
With option -c shadow the second and all subsequent morphologies
are plotted as underlying structures with the first morphology on top of
them. The default shadow color is lightgray and the line width 3.0. To
make a plot as in the figure above, issue the following command:
swc view pass_nmo_2_cut.swc rep.swc -p 3 -c shadow --shadow-color red --shadow-width 0.5
Modifying morphologies
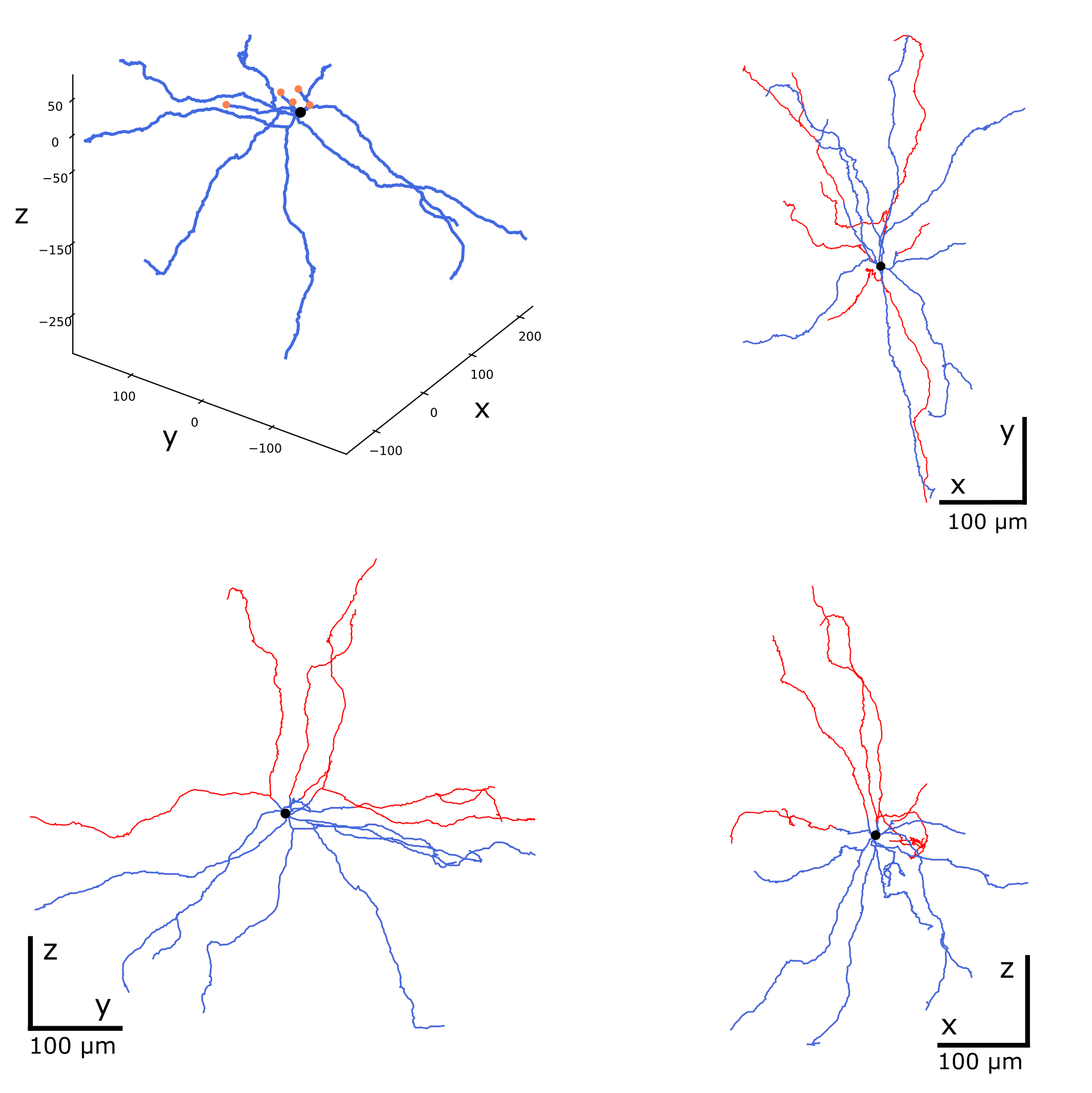
Correct or repaired morphology reconstructions may need further modifications before entering the simulation pipeline. A typical example is cloning of the finished reconstructions with random manipulation of their neurites. This increases variability within the population of morphologies keeping its topological structure and statistical features, as shown in the figure. See the corresponding options of the commands modify and repair.
Morphology modifications are used for cloning the available reconstructions to achieve higher morphological variability in the simulations. Let’s take as an example the random morphology created by the command repair as in the section above:
swc repair pass_nmo_2_cut.swc -c `swc find pass_nmo_2_cut.swc -c 10 -p 3` --seed 123
We modify repaired morphology rep.swc by twisting dendrites at the
branching points to a random angle up to 360 degrees and then scale the
resulting morphology mod.swc in (x, y, z) by the factor of 0.8. The
result is saved into clone1.swc:
swc modify rep.swc -p 3 -w 360
swc modify mod.swc -s 0.8 0.8 0.8 -o clone1.swc
Likewise, we twist dendrites of the reconstruction rep.swc and scale
it by 1.2, creating morphology clone2.swc:
swc modify rep.swc -p 3 -w 360
swc modify mod.swc -s 1.2 1.2 1.2 -o clone2.swc
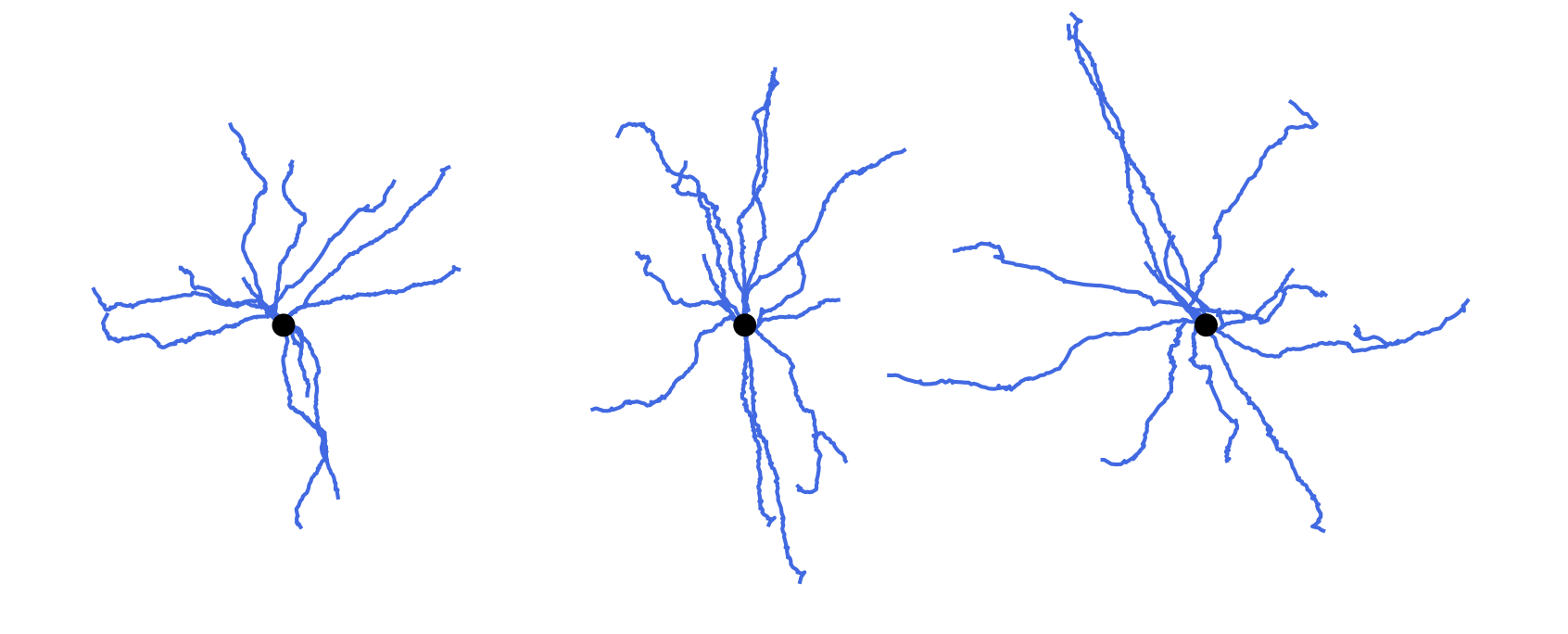
Cloning morphologies with random modifications. Original morphology in the middle. Cloned morphologies have the dendrites twisted randomly at the branching points and scaled by the factor of 0.8 (on the left) and 1.2 (on the right).
Finally, ready reconstructions can be resampled with fixed spatial resolution. This operation preserves position of the structure-defining points (i.e. neurite stem points, branching points and terminals) but slightly reduces the total length.
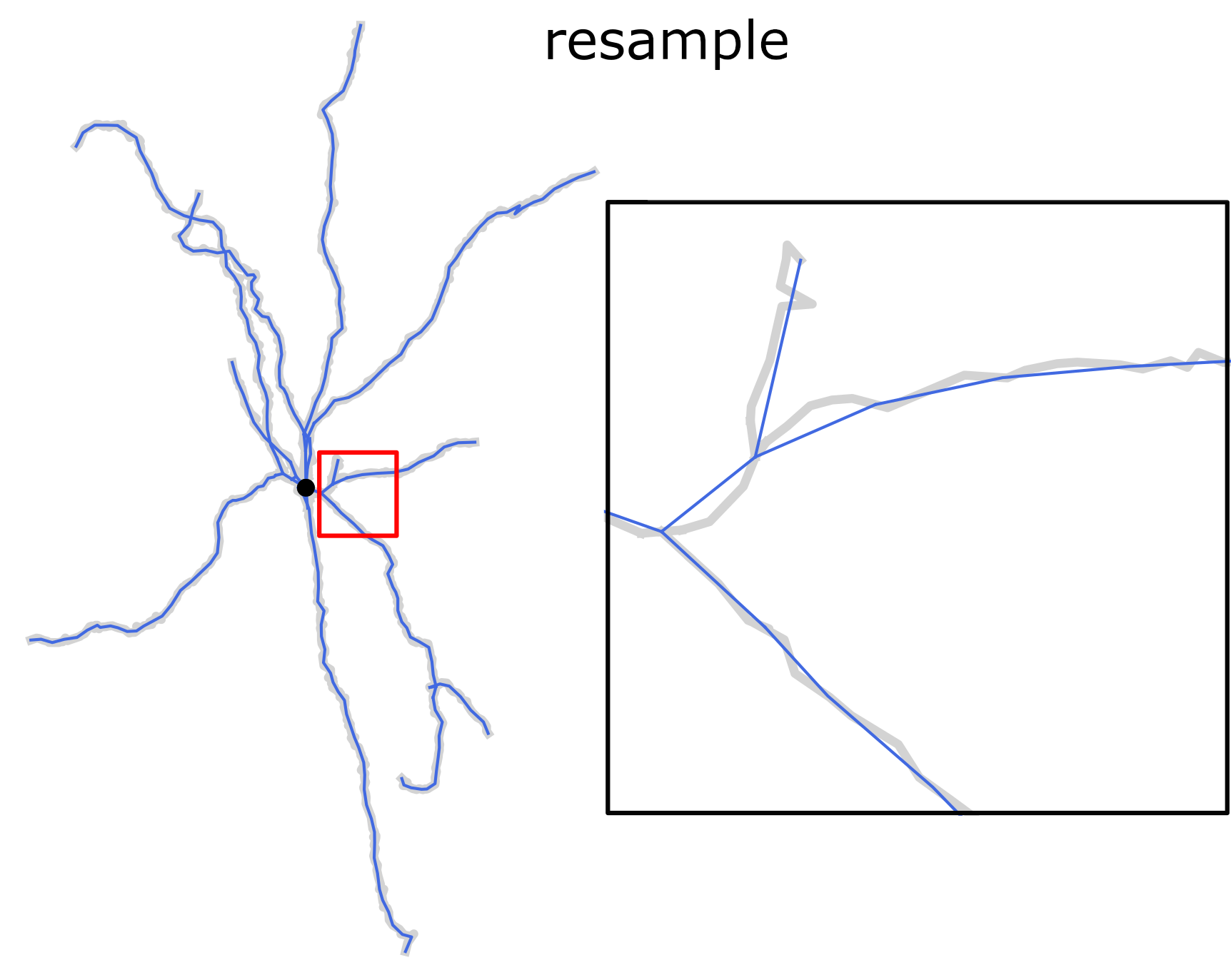
Resampling morphology reconstruction with fixed spatial resolution. Structure points-preserving operation.
For the full list of available options provided by module treem,
see Command-line interface and API reference.